Preferences
Choosing Favorites... Preferences from the menu opens the
Preferences Tool.
Options within are grouped by Category:
By default, preferences are stored in the file
$HOME/.chimera/preferences, where $HOME refers to
the user's home directory. The location can be changed in the
Preferences section
of the Preferences Tool.
The preferences file can contain information in addition to
the Preferences Tool settings,
including the names of recently accessed files and directories.
For just the category being shown:
Reset replaces the current settings with the original defaults,
Restore replaces the current settings with
those previously saved in the
preferences file,
and Save saves the current settings to the
preferences file.
Equivalent operations can be performed for
all categories at once
within the Preferences section
of the Preferences Tool.
Help opens this manual page in a browser window, and
Close dismisses the Preferences Tool.
Default settings are indicated below in bold.
Background
The Background section of the
Preferences Tool includes:
- Background color (a
color well, No Color by default)
- when set to no color, the background is black
- Label font (Sans Serif/Serif/Fixed)
(Normal/Bold/Italic/Bold Italic) (6/8/9/10/11/12/16/18/24/30/36)
- Selection highlight method (outline/fill) - how
a selection is highlighted in the graphics window
- Selection highlight color (a
color well, green by default) -
what color to use for highlighting a selection
in the graphics window
- Draw lens borders (true/false) - whether to draw outlines
of any lenses in the graphics window
- Lens border color (a
color well, green by default)
- what color to use for outlining any
lenses in the graphics window
Collaboratory
The Collaboratory section of the
Preferences Tool controls some aspects
of the
Collaboratory extension. Setting a port number and publishing
the IP address may help enable use under certain
problematic
network conditions.
- Logging (not yet implemented)
- Collaboratory logging enabled (true/false) - whether
to save Collaboratory messages in a log file
- Collaboratory log location - pathname for the log file
- IP address
- Publish IP enabled (true/false) - whether to override
the IP address supplied by the computer with the address specified below;
needed when a computer supplies a "local" IP address rather than the
address needed for internet communications
- IP address to publish - the IP address of the computer or
router supplying the internet connection
- Port number
- Publish port enabled (true/false) - whether to designate
a specific network port number for communications
(otherwise, the port will be randomly allocated by CORBA)
- Port to publish - the port number (must be greater than 1024,
as ports up to 1024 are reserved for use by the system)
General
The General section of the
Preferences Tool includes:
- Confirm exit (true/false) - whether to ask for
confirmation after Session... Exit has been chosen from the menu
- Show balloon help (true/false) - whether to show
balloon help,
explanatory text that pops up when the cursor is left in certain positions
- Show atomspec balloon
(true/false) - whether to show
label information when the cursor is left on top of an atom or bond.
- Atomspec display style
(simple/command-line specifier/command-line-dependent) -
whether to describe atoms with a simple style that consists of residue
name, residue number, and atom name, or with an
atom specification string
(affects atomspec balloons and the
Structure Measurements panels); the default is to use
atom specification strings
when the Command Line
is shown and the simple style when the
Command Line is not shown
- File lists path style (file - leading path/full path) -
how to list files and their locations in certain dialogs (such as
Files to read at startup within the Midas
section of the Preferences Tool); a change
will not be evident until after Chimera is restarted
- Toolbar visible (no/yes/auto) - whether or not to display
the toolbar,
where icons are placed; the auto option
only shows the toolbar if it contains one or more icons
- Toolbar side (left/right/top/bottom) - position of the
toolbar relative to the graphics window
- Use short open/save browser (true/false) - whether
open/save dialogs
should show only the two lowest directory levels
(more details)
- Open dialog starts in directory from last session
(true/false) - whether the first
open/save dialog invoked during a session should start in the directory
where a file was last opened or saved in the previous session, or in the
current working directory. The default is true on
Windows systems, false on the others.
Image Credits
The Image Credits section of the
Preferences Tool specifies information to be included
in saved image files (TIFF, PNG).
This information does not affect the image itself.
- Artist (user's login name by default) - optional, name of the
image creator
- Copyright (blank by default) - optional, a copyright statement
Image Setup
The Image Setup section of the
Preferences Tool sets specifications for images
saved to files using the Image Panel.
This section can also be invoked from the
Image Panel.
- Units
(centimeters/inches/millimeters/points)
- the units used for describing the options below
- Image Resolution
- Pixels per unit (by default, 100 per inch or
the equivalent in other units) - the desired resolution;
multiplied by the image width and height in the
Image Panel to yield the dimensions in pixels
- Resolution Calculator - suggests a resolution that minimizes
output file size for printing purposes by computing the optimal number
of printer pixels required to represent one screen pixel
- Printer dots per unit (300 by default)
- Colors per dot (3/3+black/4/5/5+black/6)
- Shades per color (2 by default)
- Antialiasing - used to decrease jaggedness along straight edges
within saved images (does not affect the interactive display)
- Supersample (1x1/2x2/3x3/4x4/5x5/6x6)
- how many pixels are sampled in the X and Y dimensions for each pixel in
the final saved image; higher values increase smoothness and calculation time
Messages
The Messages section of the
Preferences Tool controls the disposition of
command, status, warning, and error messages.
- Show status line (true/false) - whether to
show a line for status messages under the main
Chimera window (otherwise, status messages are not visible);
can also be set from the
Accelerators dialog
- Clear status line after (5 seconds/10 seconds/20 seconds/30
seconds/1 minute/never) - how long a status message will be shown in
the status line (when not overwritten by subsequent status messages);
applies to status messages appearing in certain dialogs as well as
the main Chimera status line
- Command (reply log only/dialog) - whether command messages
should appear in the Reply Log
only, or also generate a dialog box
- Warning (reply log only/dialog) - whether warnings
should appear in the Reply Log
only, or also generate a dialog box
- Error (reply log only/dialog) - whether error messages
should appear in the Reply Log
only, or also generate a dialog box
Midas
The Midas section of the
Preferences Tool is only available when the
Command Line
(Midas Emulator) is shown.
Files to read at startup refers to what
command files, if any,
should be executed when the
Command Line is started.
There are two defaults:
- $HOME/.chimera/midasrc (in a directory named .chimera in
the user's home directory, here referred to as $HOME)
- .chmidasrc in the current directory
and either or both are executed (in the order listed), if found.
The defaults can be deleted and/or additional names and locations
can be added to the list.
The files are treated as if they had been
sourced in the
Command Line.
Whether files are listed by name followed by path or with
the path preceding the name is controlled in the
General preferences.
The midasrc mechanism is similar to that in MidasPlus, where
a file named .midasrc in the user's home directory was automatically
executed upon program startup. In general, the same file can be used
here after commands unsupported by Chimera
have been removed.
Mouse
By default, structures are rotated with the left mouse button
and XY-translated with the middle mouse button (for more
details, see mouse manipulation).
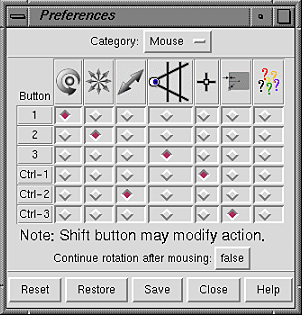 Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).
Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).
Continue rotation after mousing (true/false) indicates whether
to rotate continuously when the mouse button assigned to rotation is released
while the cursor is in motion.
New Molecules
The New Molecules section of the
Preferences Tool controls how newly opened molecules
will initially appear:
- use new color for each model (true/false) -
set the model-level color
(see coloring hierarchy)
differently for each new molecule model;
submodels with
the same main model number will be assigned the same model-level color.
This preference can also be controlled with
set/~set autocolor.
- otherwise, use color (a
color well, by default showing
a light gray color) -
color at the model level, see coloring hierarchy
- ribbon display (off/on)
- whether to show a secondary structure ribbon;
ribbons are only drawn for proteins and nucleic acids.
Protein secondary structure is determined from HELIX and SHEET records
in the input PDB file or by using
ksdssp.
- ribbon cross section (edged/flat/round/any additional
styles
created and named with the
Ribbon
Style Editor) -
style
of secondary structure ribbon shown
(see also ribrepr)
- ribbon scaling (Chimera default/any additional
scalings
created and named with the
Ribbon
Style Editor)
- secondary-structure-specific
scaling
of ribbon height and width
- ribbon hides backbone atoms (true/false) -
whether showing ribbon for residues within the model hides
backbone atoms for those residues (see the command
ribbackbone for details)
- line width (1.0 by default)
- pixel width of lines depicting bonds (when in the wire draw mode)
- atom style (dot/ball/endcap/sphere) - atom
draw mode
- bond style (wire/stick) - bond
draw mode
- stick scale (1.0 by default)
- scale factor for bonds in the stick
draw mode.
The stick scale is multiplied by individual bond radii (default
0.2 angstroms) to generate stick radii in angstroms.
Changing stick scale is preferable to changing all of the bond radii
in a model, because the former will also scale singleton atoms in the
endcap draw mode appropriately. Either way,
the other endcap atoms (those participating in bonds) will be scaled
to match the thickest of the attached bonds.
- ball scale (0.25 by default)
- scale factor for atoms in the ball
draw mode.
The ball scale is multiplied by individual atom
VDW radii to generate ball radii in angstroms.
- coloring by element (true/false) - whether
byatom coloring is used;
this overrules the model-level color above, see
coloring hierarchy
- auto-chaining (off/on)
- whether to connect atoms that precede and follow undisplayed segments
(whether to draw pseudobonds between them)
- metal complex line width (1.0 by default)
- pixel width of pseudobonds
depicting metal coordination (when in the wire draw mode)
PDB
The PDB section of the
Preferences Tool includes:
- Fetch from web as necessary (true/false) -
whether to retrieve an input PDB file from the
Protein Data Bank web site
when it is not found locally (see the Web Access
preferences for proxy settings, if needed)
- Personal PDB directories - additional directories to look
in for PDB files; the user may Add a directory to the list of
places to look, or choose a previously added directory from the list
and then Delete it from
the list
Preferences
The Preferences section of the
Preferences Tool includes:
- Preferences file - where the
preferences file
for the current session should be located:
- preferences (in the current directory)
- .chimera/preferences (in a directory named .chimera in the current
directory
- $HOME/.chimera/preferences (in a directory named .chimera in
the user's home directory, here referred to as $HOME)
- All categories:
Tools
The Tools section of the
Preferences Tool allows control over the following
Settings for each tool (extension):
- Auto Start - whether to launch the tool automatically
upon Chimera startup
- On Toolbar - whether to show the tool's icon in the
toolbar
- In Favorites - whether to include the tool in the
Favorites menu
- Confirm on Start - whether to ask
for confirmation before starting the tool
The Locations section allows users to
specify additional directories for Chimera to look in for tools.
Clicking Add opens a dialog for directory specification.
Delete removes a directory from the list.
Web Access
The Web Access section of the
Preferences Tool includes:
- Confirm open of commands or code (once per session/each
time/never) - whether/how often to
ask for
confirmation before opening certain types of
data
linked to web pages. The default is to ask once per Chimera startup.
- Accept web data (true/false) - whether the running
instance of Chimera can be used as a helper application for
data
linked to web pages (and can receive files specified with
chimera --send).
On Mac OS X, this preference is not shown, and is effectively always
true. After a browser has been
configured
to send Chimera certain types of files, setting
Accept web data to true enables the running instance of
Chimera to accept such files.
If there is no running instance of Chimera enabled to accept web data,
another instance of Chimera will be started and used to open the data.
If there are multiple running instances of Chimera set to
accept web data, the file will be sent to the instance that
most recently had focus (was most recently clicked into).
- Use HTTP proxy (true/false) - whether to use
a proxy to connect to the internet through a firewall
(for example, to fetch a PDB file from the
Protein Data Bank web site);
the server and port number settings (below) should generally be the same as
those used for web browsers
- Proxy server - the hostname or IP address of the proxy server
- Proxy port - proxy port number to use
 Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).
Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).