aMatReader: A Cytoscape Reader for Adjacency Matrices
aMatReader is a Cytoscape 3 app that augments the Cytoscape network readers by adding the ability to read in an adjacency matrix. The input file must have the ".mat" extension to avoid conflict with other readers and the format of the file should have the names of the nodes in the top for and as the first field in each row thereafter. Note that the first field of the first (header) row is ignored. Each of the values in the fields are interpreted as edge weights. Missing data is assumed to indicate that no edge exists between two nodes. There is no assumption that the nodes defined in the header row and the nodes defined in each data row are in the same order or are matching.
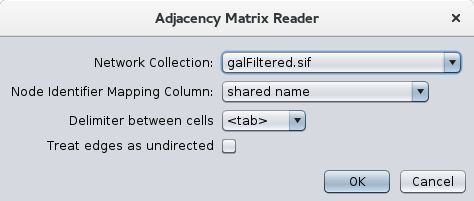
Once the App is installed from the app store, using it is a simple matter of doing a File→Import→Network→File and choosing a file with a ".mat" extension. Once the file has been chosen, the dialog shown in Figure 1 will be displayed. The top part of the dialog (Network Collection and Node Identifier Mapping Column) are standard for importing files when there is already a network connection. These two items will not show if there are currently no network collections. The next two items allow you to choose the delimiter for your adjacency matrix (currently supported are tab, comma, vertical bar, and space) and whether or not to treat the edges as undirected. If you choose undirected edges, only one edge will be imported for each pair of nodes. In some cases, it might be better to leave this unchecked and collapse duplicate edges later.
Sample File: sample.mat
#Simple tab-delimited adjacency matrix
Node 1 Node 2 Node 3 Node 4 Node 1 1 4 Node 2 1 3 3 Node 3 4 3 1 Node 4 3 1
| [Contents] [Top] |
Last updated on November 25, 2015
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2021 Regents of the University of California. All rights reserved.