See also: ChimeraX Toolshed
AFMFormatThe AFMFormat bundle for ChimeraX enables reading atomic force microscopy data in the .afm file format, as described in Jiang, Wang, and Scheuring, Nat Commun 16:1671 (2025). The paper's supplementary information includes installation instructions. Without the bundle, however, such files can already be read if treated as MRC format.ANNOTEX
ANNOTEX (Annotation Extension for ChimeraX) facilitates manual curation of functional annotations based on top hits from sequence searches with Diamond, structure searches with Foldseek using ColabFold predictions as queries, eggNOG functional annotations, and DeepTMHMM predictions of TMD and signal-peptide segments. The method is described in Svedberg et al., BMC Genomics 25:6 (2024).ATOMDANCE
ATOMDANCE performs machine-learning-assisted comparison of molecular dynamics of a protein in two different functional states, such as bound vs. unbound or mutant vs. wild-type. It highlights the residues with the most differences and interfaces automatically with ChimeraX to produce color-mapped images and movies. ATOMDANCE is available on github and described in Babbitt et al., Biophys J 123:2705 (2024).AutoEMage
AutoEMage automates cryo-electron (cryo-EM) microscopy file transfer, motion correction, CTF estimation, image display, outlier detection, particle picking, 2D classification, class ranking, and model generation. It uses MotionCor2, CTFFIND4, UCSF ChimeraX, RELION, and IMOD, and is described in Cheng et al., J Appl Cryst 56:1865 (2023).ContinuousFlex
ContinuousFlex is a plugin to Scipion for continuous conformational flexibility analysis of cryo-EM data. It uses ChimeraX for fitting and visualization, as described in Vuillemot et al., Int J Mol Sci 25:20 (2023).DeepSSETracer
DeepSSETracer uses a trained CNN model to detect helices and β-sheets from a cryo-EM map of medium resolution (5-10 Å). It is integrated with ChimeraX for visualization, as described in Mu et al., Bioinform Adv 4:vbae169 (2024).DomainFit
DomainFit is a program for automated domain-level protein identification from cryo-EM maps at resolutions > 4 Å. It uses ChimeraX to fit AI-predicted domain structures (such as from AlphaFold) into the maps and cluster the fits, and then it performs statistical analyses to try to identify which proteins form the density. See the example and the paper by Gao et al. (preprint 2023, DOI: 10.1101/2023.11.28.569001).MitoTNT
MitoTNT builds on MitoGraph segmentation and ChimeraX visualization for the analysis of mitochondrial networks in 4D live-cell fluorescence microscopy data, as described in Wang et al., PLoS Comput Biol:e101160 (2023).MorphOT
MorphOT morphs density maps using shape interpolation based on optimal transport, as described in Ecoffet et al., Bioinformatics:btaa1019 (2020).PTM-Psi
PTM-Psi is a Python package that includes a ChimeraX plugin to replace amino acid sidechains with certain post-translationally modified (PTM) forms, as described in Mejia-Rodriquez et al., Protein Sci:e4822 (2023). The initial library includes several common cysteine PTMs, and a broader range of types may be offered in the future.pyCapsid
pyCapsid is a Python package to extract quasi-rigid domains in protein shells and other macromolecular complexes, with output including a script for ChimeraX visualization. It is available as a Google Colab notebook and as code on Github for those who wish to install it locally. See the online tutorial and Brown et al., Bioinformatics 40:btad761 (2024).pyLattice
pyLattice is a library of Python modules, Jupyter notebooks, and Matlab routines that operate together to form an image analysis pipeline for lattice light-sheet microscopy, as described in Schöneberg et al., Mol Biol Cell 29:2959 (2018). It provides customized input for 4D (3D + time) visualization in ChimeraX.subtomo2Chimera
Python scripts to convert Dynamo table and relion 4.0 (and 3.1) star file to ChimeraX cxc script to visualize the subtomograms within the tomogram.RNArtist
RNArtist allows designing RNA 2D structures interactively and using UCSF ChimeraX (or UCSF Chimera) to explore 3D architectures. RNArtist is developed by Fabrice Jossinet at the Institut de Biologie Moléculaire et Cellulaire, Strasbourg.
 |
The ScanNet server predicts protein-binding sites for
structures in the PDB, the AlphaFold Database, or uploaded from a local file.
The method is described in
Tubiana et al.,
Nat Methods 19:730 (2022)
and the web server specifically in
Tubiana et al.,
J Mol Biol 434:167758 (2022).
Results are emailed to the user as a zip file containing:
|
| SymProFold combines AlphaFold-Multimer predictions with general symmetry considerations to predict symmetric protein assemblies such as bacterial S-layers and viral capsids. It creates an mmCIF file containing the primitive unit cell and symmetry information of the predicted assembly. The related tool Domain_Separator separates an atomic structure into structural domains. Both tools use ChimeraX methods and libraries and are described in Buhlheller et al., Nat Commun 15:8152 (2024). |
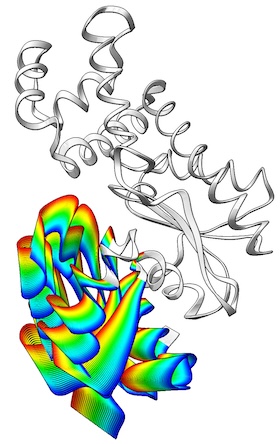 |
The ViewMotions Server
generates a representation of a protein conformational change in a visually
intuitive way using a color spectrum to indicate the molecular motion.
The method and server are described in
Cockrell & Kantrowitz,
J Mol Graph Model 40:48-53 (2013).
The two input structures can be specified by PDB identifier or supplied
as two PDB files to be uploaded to the server.
Results are e-mailed to the user as a tar file containing a PNG image
and scripts for visualizing the results in:
|
Wiggle is a ChimeraX plugin for visualizing conformational heterogeneity within cryo-EM data, as calculated by cryoDRGN or by cryoSPARC 3D variability analysis; see the user interface overview and other video tutorials. The cryoDRGN and cryoSPARC analyses depend on a CUDA-accelerated GPU, and Wiggle is currently experimental (hence not yet available on the ChimeraX Toolshed).xiView
xiView is a web-based visualisation tool for the analysis of cross-linking mass spectrometry results, as described in Combe et al., J Mol Biol 436:168656 (2024). 3D export options include UCSF ChimeraX pseudobond files.