Movie Making in Chimera
Tom Goddard
May 5, 2009
UCSF Mission Bay Library
Goals
- Make simple molecular movies.
- Get best movie quality.
- Learn Chimera commands that are useful for movie making.
Scripts and PDB files used: movie_data.zip
Spin 360 degrees - The mother of molecular animations
Open maltotriose binding protein and make it look good.
File / Open -> 2gha-chain-A.pdb
Presets / Interactive 1 (ribbons)
Spin and make movie.
Favorites / Command-line
turn y 3 120
Tools / Utilities / Movie Recorder
Record, turn y 3 120, Stop
Output file Browse, Desktop/m1.mov, Make Movie
double click m1.mov on desktop and play
Note: default playback speed is 25 frames per second.
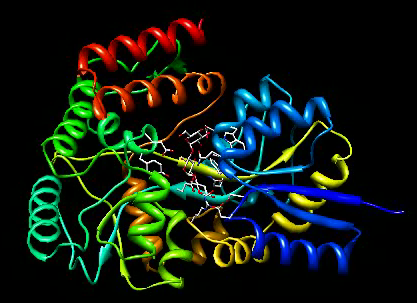 Cut out initial/final segments with no motion.
movie record ; turn y 3 120 ; wait 120 ; movie stop
movie encode output ~/Desktop/m2.mov
Play movie m2.mov
Rock instead of spin to emphasize bound ligand.
rock y
freeze
rock y 3 68 # 3 degree steps, 68 frames for one cycle
rock x 3 68
File / Open script m3.cmd to record movie
movie record
rock y 3 68
wait
rock x 3 68
wait
movie stop
movie encode output ~/Desktop/m3.mov
File / Open -> m3.cmd
Play movie m3.mov
Improving the movie appearance. Spin centered on the maltriose ligand.
Cut out initial/final segments with no motion.
movie record ; turn y 3 120 ; wait 120 ; movie stop
movie encode output ~/Desktop/m2.mov
Play movie m2.mov
Rock instead of spin to emphasize bound ligand.
rock y
freeze
rock y 3 68 # 3 degree steps, 68 frames for one cycle
rock x 3 68
File / Open script m3.cmd to record movie
movie record
rock y 3 68
wait
rock x 3 68
wait
movie stop
movie encode output ~/Desktop/m3.mov
File / Open -> m3.cmd
Play movie m3.mov
Improving the movie appearance. Spin centered on the maltriose ligand.
 Select / Residue / MLR, then Actions / Set Pivot
windowsize 640 480 # Larger window size
set bg_color white # white background
Tools / Viewing Controls / Effects, silhouettes on
Open script m4.cmd to record movie.
Play movie m4.mov
Improve rendering quality. Increase movie bitrate.
Use "movie encode bitrate 10000" in script m5.cmd, default bitrate 2000 Kbits / sec.
Movie file size will increase proportional to bitrate.
Play movie m5.mov
Select / Residue / MLR, then Actions / Set Pivot
windowsize 640 480 # Larger window size
set bg_color white # white background
Tools / Viewing Controls / Effects, silhouettes on
Open script m4.cmd to record movie.
Play movie m4.mov
Improve rendering quality. Increase movie bitrate.
Use "movie encode bitrate 10000" in script m5.cmd, default bitrate 2000 Kbits / sec.
Movie file size will increase proportional to bitrate.
Play movie m5.mov

| 
|
| Default bitrate 2000.
| Bitrate 10000 Kbits/sec.
|
To avoid final frame of movie looking bad, add 10 stationary frames at end.
Add "wait 10" command before "movie stop", m6.cmd
Play movie m6.mov
Reduce jagged edges. Use supersampling.
Use "movie record supersample 3" in script m7.cmd
9 times slower recording movie.
Play movie m7.mov
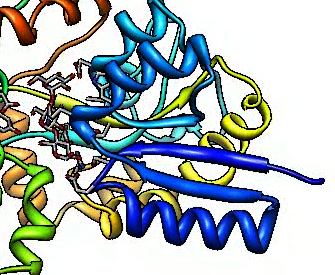
| 
|
| No supersampling.
| Supersample 3 by 3.
|
Raytraced rendering to produce shadows.
Use "movie record raytrace true" in script m8.cmd
Very slow recording movie. Time depends on scene complexity and window size.
Make window size small and only show ligand as spheres for example.
Play movie m8.mov

Volume data movie
Movie showing sections of rat cortex from electron microscopy.
File / Close session
File / Open -> ratbrain.cmap
Volume dialog, style solid, Features / Planes, One plane
windowsize 892 772 # match volume data size
Move volume dialog Plane slider.
volume #0 planes z,0,100
Open script r1.cmd to record movie.
Play movie r1.mov

Command Documentation
Two easy ways to access Chimera command documentation. Use "help" command
help volume
Or go to Chimera web site (google Chimera), click Command Index link at left.
Documentation for the movie command links to other useful commands.
Commands do not exist for all Chimera capabilities (for example, Morph Map).
Animation gallery at Chimera web site has example animations.
More Movie Examples
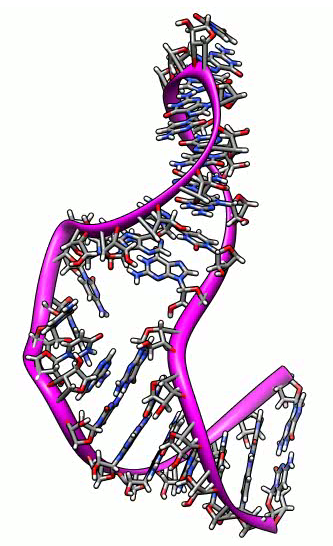
Morphing between atomic models. NMR ensemble of RNA stem loop that convinces
ribosomes to translate viral RNA without methylguanosine mRNA cap.
Molecular dynamics trajectories. Beta hairpin of G protein with hydrogen bonds
identified at each frame of trajectory.

Morphing between two maps. RNA polymerase in two conformations.
More examples from Chimera
animation gallery.
Other Animation Software
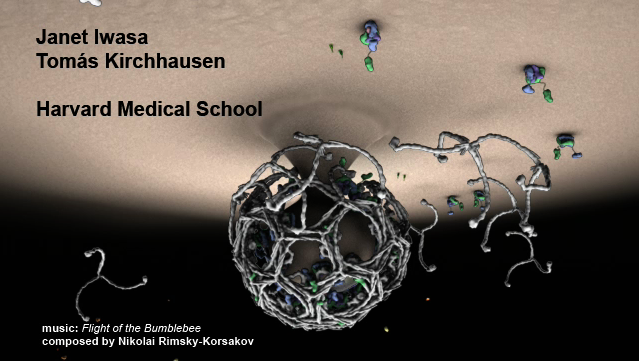
Chimera makes only simple animations. Here is a movie made using
Autodesk Maya commercial animation software with data exported from Chimera.
Clathrin cage assembly by Janet Iwasa, Harvard Medical School.
Start playing mid-way through.
Chimera Animations with Multiple Stages
Example Chimera movie with several stages.

Movie mbp.mov made with script mbp.cmd.
How to add titling.
Tools / Utilities / 2D Labels, click in graphics window and type text.
Move label with mouse.
Command version 2dlabel allows fading labels.
(Dialog label ids are label2d_id_0, label2d_id_1, ...)
2dlabel create MBPintro text "Binding maltotriose" xpos 0.3 ypos 0.92 color black
2dlabel change MBPintro visibility hide frames 25
(Chimera 1.4 bug makes fade abrupt after solid volume shown. Restart.)
Making the maltotriose ligand fly.
File / Close Session
File / Open, 2gha-chain-A.pdb and 2ghb-chain-A.pdb
Presets / Interactive 1 (ribbon)
Select / Residue / MLR
Select / Invert (selected models)
Actions / Ribbon / hide
Actions / Atom+Bond / hide
Move models to lower left corner
savepos p1
Active models, turn off #1 with checkbutton below command-line.
Move ligand to upper right with mouse and rotate.
savepos p2
reset p1 100
Command script f1.cmd for flying ligand f1.mov.

Exercise: Morph Molecule
Use the morph conformations tool to show different configurations of
the HIV packaging signal, RNA stem loop 1 (pdb 1m5l).
File / Close session
File / Open, 1m5l.pdb
Presets / Interactive 1 (ribbon)
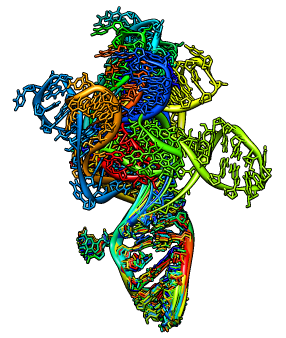 Tools / Structure Comparison / Morph Conformations
Morph dialog, Add... button, drag mouse to select
all 15 models, press Add.
Then choose first model, and press Add.
This makes morph loop back to starting conformation.
Morph dialog, set Action on Create to "hide conformations".
Morph dialog, press Create.
Actions / Color / by element
MD Movie dialog will appear, press play button (triangle pointing right).
MD Movie dialog, File / Record movie, /Users/.../Desktop/1m5l.mov, Record
Command script 1m5l.cmd for RNA morph 1m5l.mov.
Tools / Structure Comparison / Morph Conformations
Morph dialog, Add... button, drag mouse to select
all 15 models, press Add.
Then choose first model, and press Add.
This makes morph loop back to starting conformation.
Morph dialog, set Action on Create to "hide conformations".
Morph dialog, press Create.
Actions / Color / by element
MD Movie dialog will appear, press play button (triangle pointing right).
MD Movie dialog, File / Record movie, /Users/.../Desktop/1m5l.mov, Record
Command script 1m5l.cmd for RNA morph 1m5l.mov.
 Cut out initial/final segments with no motion.
movie record ; turn y 3 120 ; wait 120 ; movie stop
movie encode output ~/Desktop/m2.mov
Play movie m2.mov
Rock instead of spin to emphasize bound ligand.
rock y
freeze
rock y 3 68 # 3 degree steps, 68 frames for one cycle
rock x 3 68
File / Open script m3.cmd to record movie
movie record
rock y 3 68
wait
rock x 3 68
wait
movie stop
movie encode output ~/Desktop/m3.mov
File / Open -> m3.cmd
Play movie m3.mov
Improving the movie appearance. Spin centered on the maltriose ligand.
Cut out initial/final segments with no motion.
movie record ; turn y 3 120 ; wait 120 ; movie stop
movie encode output ~/Desktop/m2.mov
Play movie m2.mov
Rock instead of spin to emphasize bound ligand.
rock y
freeze
rock y 3 68 # 3 degree steps, 68 frames for one cycle
rock x 3 68
File / Open script m3.cmd to record movie
movie record
rock y 3 68
wait
rock x 3 68
wait
movie stop
movie encode output ~/Desktop/m3.mov
File / Open -> m3.cmd
Play movie m3.mov
Improving the movie appearance. Spin centered on the maltriose ligand.
 Select / Residue / MLR, then Actions / Set Pivot
windowsize 640 480 # Larger window size
set bg_color white # white background
Tools / Viewing Controls / Effects, silhouettes on
Open script m4.cmd to record movie.
Play movie m4.mov
Improve rendering quality. Increase movie bitrate.
Use "movie encode bitrate 10000" in script m5.cmd, default bitrate 2000 Kbits / sec.
Movie file size will increase proportional to bitrate.
Play movie m5.mov
Select / Residue / MLR, then Actions / Set Pivot
windowsize 640 480 # Larger window size
set bg_color white # white background
Tools / Viewing Controls / Effects, silhouettes on
Open script m4.cmd to record movie.
Play movie m4.mov
Improve rendering quality. Increase movie bitrate.
Use "movie encode bitrate 10000" in script m5.cmd, default bitrate 2000 Kbits / sec.
Movie file size will increase proportional to bitrate.
Play movie m5.mov












 Tools / Structure Comparison / Morph Conformations
Morph dialog, Add... button, drag mouse to select
all 15 models, press Add.
Then choose first model, and press Add.
This makes morph loop back to starting conformation.
Morph dialog, set Action on Create to "hide conformations".
Morph dialog, press Create.
Actions / Color / by element
MD Movie dialog will appear, press play button (triangle pointing right).
MD Movie dialog, File / Record movie, /Users/.../Desktop/1m5l.mov, Record
Command script 1m5l.cmd for RNA morph 1m5l.mov.
Tools / Structure Comparison / Morph Conformations
Morph dialog, Add... button, drag mouse to select
all 15 models, press Add.
Then choose first model, and press Add.
This makes morph loop back to starting conformation.
Morph dialog, set Action on Create to "hide conformations".
Morph dialog, press Create.
Actions / Color / by element
MD Movie dialog will appear, press play button (triangle pointing right).
MD Movie dialog, File / Record movie, /Users/.../Desktop/1m5l.mov, Record
Command script 1m5l.cmd for RNA morph 1m5l.mov.