

MSF Viewer allows viewing and manipulation of multiple sequence alignments in MSF format. If the corresponding structures are displayed in Chimera, there can be crosstalk showing which regions on the structure are those highlighted in the sequence, and vice versa. The alignment may be edited, highlighted in various regions, and written back out in MSF format or in color PostScript for presentation or publication.
MinRMS, a stand-alone program available from the UCSF Computer Graphics Lab, produces structural superpositions and other data that can be displayed with AlignPlot, as well as sequence alignments that can be displayed with MSF Viewer (and Multalign Viewer). However, many other programs can generate MSF files suitable as input to MSF Viewer. Likewise, structural alignments can be generated in a multitude of ways.
MSF Viewer is no longer under development; Multalign Viewer is generally more advanced and preferred for working with sequence alignments.
There are several ways to start MSF Viewer, a tool in the Homology category. Explicitly starting MSF Viewer opens a dialog box requesting the name of the input MSF file.
Once the file has been read, the sequence window containing the alignment appears:
| MSF Viewer sequence window |
|---|
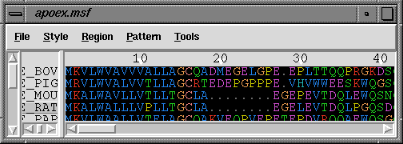
|
To allow the proper correspondence between sequences and structures, the names of the sequences in the MSF file should match the filenames of structures opened in Chimera. Additional sequences which do not match any structure and additional structures which do not match any of the sequences are permitted; it is even possible to use MSF Viewer without opening any structures. If the name of a structure file does not match the name of the corresponding sequence in the MSF file, the two can be linked by positioning the cursor over the sequence name in the sequence window, pressing the middle mouse button, and selecting Associate PDB... (if the structure has already been opened) or Associate New PDB... (to open and then associate the structure with the sequence). This pop-up menu also includes Inspect... (same thing as File... Inspect in the sequence window menu), Disassociate PDB... (close the associated model), and Set Model Color...
Regions of interest in the sequence alignment can be created using the left mouse button. A box will be drawn around the residues, and the corresponding parts of the structures in the main Chimera window will be highlighted in some way. The default type of highlighting is selection; with the Region Browser (Region... Region Browser), one can specify a stick representation instead of selection, or Normal, meaning whatever the representation would be otherwise (usually the same as the rest of the molecule). Multiple discontinuous sequence regions can be chosen; the Shift key is used to add to an existing choice instead of replacing it. To reverse the flow of information and instead use selection in the main window to determine which residues are boxed in the sequence window, turn on Tools... Monitor Selection.
The active region is the latest region created, or the region whose description is chosen (by clicking) in the Region Browser; the active region is distinguishable by a border thicker than that of other regions in the sequence window. If the borders in the sequence window are not visible, changing the background color (Style... Change Background) or border color (by clicking on the corresponding color bar in the Region Browser) should help.
Another way to define regions is to read a seqsel file generated by the program JEvTrace. Region... Import Seqsel File reads a seqsel file and colors residues in the multiple alignment and in any associated structures accordingly. The seqsel file format is very simple, so users wishing to color residues alike in sequence and structure (for any reason) may wish to create a file manually and read it in by this route.
Finally, regions are created as side effects of using Style... Percent Identity and Style... Secondary Structure (described in more detail under COLORS AND FONTS). Regions defined by secondary structure are shown in the Normal representation in any associated structures, regardless of the setting within the Region Browser.
The sequence alignment in the MSF file can be used to create a structural alignment of proteins associated with the sequences. Tools... Match Aligned Residues opens a dialog requesting a Reference Structure and a Structure to Align. There can be only one reference structure, but there can be more than one structure to align. The alpha-carbons of residue pairs aligned within the MSF file are fit by a least-squares method. Each structure to align is moved to superimpose with the reference structure, which is not moved during the match.
The Style section of the sequence window menu controls the overall appearance of the window's contents. The background color of the window can be altered with Change Background. The mapping of colors to one-letter amino acid codes is shown when Edit Color Map is chosen. The map can be edited and then saved with Save Color Map for use in subsequent sessions (Style... Load Color Map). Various built-in color maps can be accessed with Change Color Scheme. Several fonts are available (Style... Change Font Family). Revert on the font panel will not undo changes; it will only revert the panel to its default state if the Apply button has not yet been clicked. Of the changes available under Style..., only changes in the mapping of colors to residue codes are applied to any PostScript outputs, which have a white background overall and use a standard font.
Style... Percent Identity brings up a dialog box for coloring the backgrounds of positions with 100% identity and positions with high percent identity (the threshold is user-specified). Each position that falls into either category becomes a region. The colors can be changed within a special color editor that appears when Set... is clicked.
Style... Secondary Structure brings up a dialog box for coloring backgrounds according to secondary structure. For protein(s) with 3D structure(s) opened in Chimera, secondary structure is determined from HELIX, SHEET, and TURN records in the input file, or if these are not present, using ksdssp. For the other proteins (present as sequence only), helix and strand segments are predicted using GOR. Contiguous segments assigned the same type of secondary structure within a sequence become regions. Clicking one of the four bars of color (for helix, sheet, and turn backgrounds, plus the border around segments determined by secondary structure records associated with an actual structure) invokes a special color editor in which the color can be changed.
The Region section of the sequence window menu controls the appearance of regions of interest, in both the sequence window and any PostScript output. Individual regions can be activated and then changed using the Region Browser (Region... Region Browser). Changes are always applied to the current active region, and clicking on the description of a particular region within the Region Browser makes it the active region. Only one region may be active at a time. Some of the operations available within the Region Browser are also available as separate Region menu items. Properties under control include background color, foreground color (the color of the text), whether or not the text is bold, border color (the color of the box outlining a region), and the color of the region in structures in the main Chimera window. Note that the background color of a region in any output PostScript files will be the same as its color in the sequence window (unlike the overall background color).
Besides the cosmetic changes described above, operations on an individual region include Clear, Delete, Reset, Raise, Lower, and Update (each is available as a button on the Region Browser and, except for Clear, also as a menu item under Region...). Delete deletes the active region entirely, so that if one wishes to add a new region without wiping out existing regions, it is necessary to use the Shift key. Reset deletes the contents of the region but leaves an undefined region active, so that to add a new region without wiping out existing regions, it is not necessary to use the Shift key. Clear deletes all of the regions, active and inactive.
Regions may overlap. The hierarchy of layers in the sequence window is reflected by the order of the region descriptions in the Region Browser. Raise and Lower move the active region up and down in the hierarchy, respectively. If regions are not overlapping, these two buttons have no visible effects.
Finally, Update is used to update a Pattern region. With the Pattern section of the menu, a particular occurrence or all occurrences of a specified sequence pattern can be made into a region. If the Ignore gaps option is turned on during the initial definition of the region, but later turned off (or vice versa), clicking Update allows the pattern region to be updated accordingly.
The alignment can be edited, although this will not automatically affect any structural superposition in the main window (see SEQUENCE ALIGNMENT TO STRUCTURE ALIGNMENT). Only gaps (spaces) can be added or deleted; sequence changes are not permitted.
The sequence alignment can be written out in two formats:
The overall background color in any PostScript output is always white, but the colors assigned to one-letter residue codes can be changed with Style... Edit Color Map. The background color, border color, text color, and text type (bold or not) of each boxed region can be altered using the Region section of the sequence window menu. To obtain encapsulated Postscript, change paper type to EPS within the Print dialog box before clicking the Save... button.